FRISBI

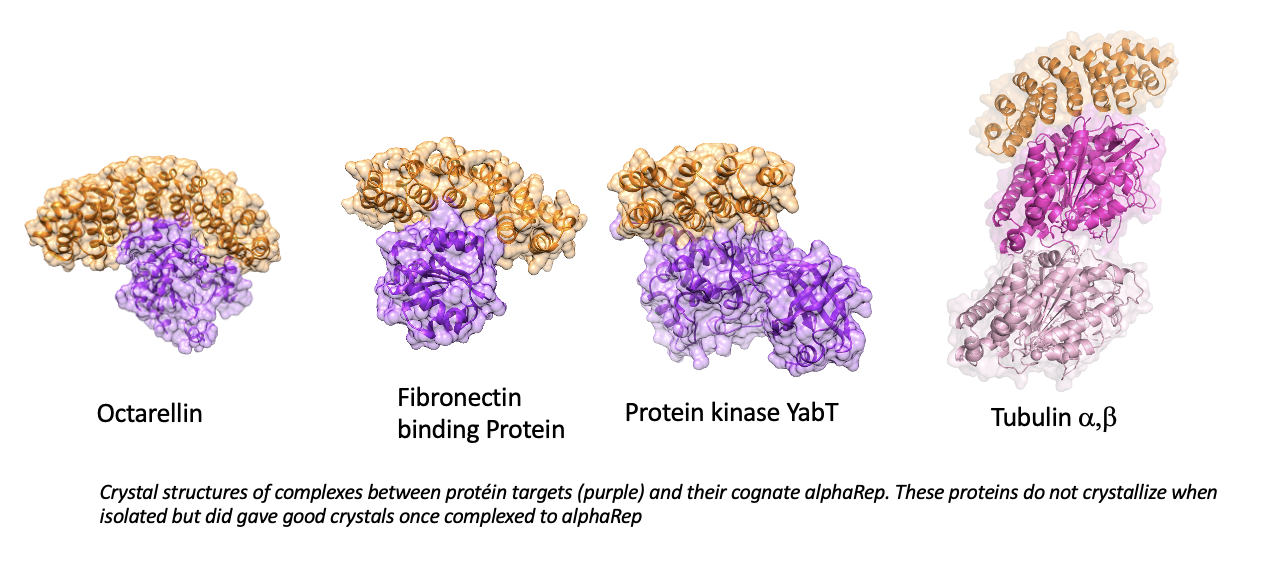
We have designed and assembled a large library of artificial repeats proteins named alphaRep (1). alphaRep proteins binding tightly and specifically any predefined folded protein can be selected out from this library (2) AlphaRep generation does not require animal immunization and gives rise to renewable, sequence defined, binders. Specific alphaReps are soluble, efficiently produced, disulfide-free and extremely stable. Upon binding, alphaRep can stabilize their specific partners, and provide a large external surface able to generate intermolecular contacts, and favor crystallization (3). Crystal structures of complexes between proteins and their cognate alphaRep have been obtained including for proteins difficult to crystallize when isolated (4). AlphaRep could also be efficiently used for electron microscopy and as specific recognition unit for cellular applications like targeted protein degradation (PROTAC) (5).