FRISBI

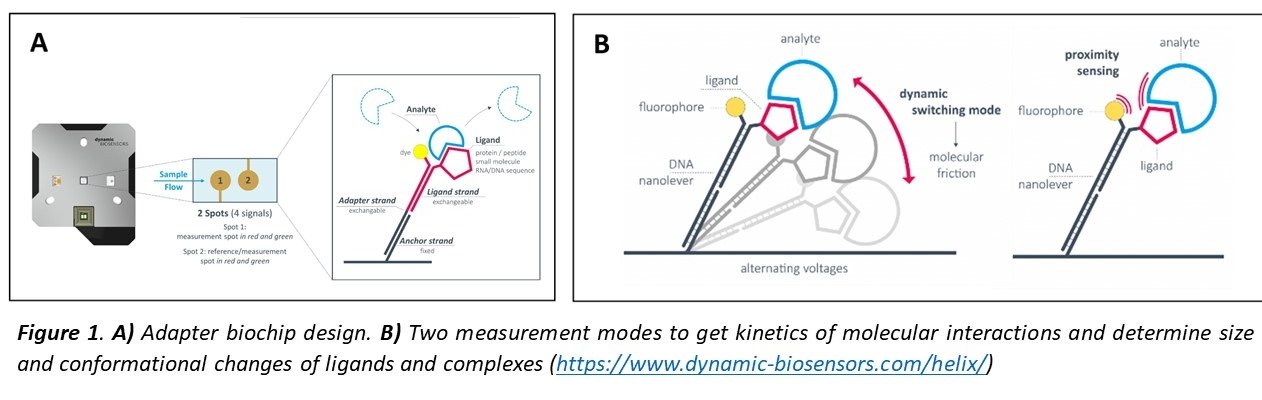
switchSENSE technology is based on short DNA nanolevers immobilized on gold electrodes in a microfluidic channel. The surface is functionalized with the ligand of interest cross-linked to the complementary DNA strands (Figure 1A). Alternating the voltage, this double strand DNA can be electrically “switched” at high frequencies1,2 and the motion tracked in real-time (µs scale) detecting a fluorescence probe (dye) present on top of one the DNA strands. Upon binding of an analyte, the hydrodynamic friction of the levers is increased and the movement slowed down, what is used to determine the size (Dh) or conformational changes of ligands and complexes. The kinetics of molecular interactions (kON, kOFF, KD) can be monitored using two measurement modes by changes in dynamic response or by Fluorescence Proximity Sensing. (Figure 1B).
The first generation DRX2 swithSENSE instrument was available since 2017 at Paris-Saclay node (I2BC, CEA Saclay site). In January 2023, it has been replaced for the new generation, heliX+, the first implemented in France. This technology has unique features such a femto-molar detention sensitivity of molecular interactions, and a confidence resolution of fastest kinetics using advanced microfluidics and 10 ms data collection. Our platform applied with success this innovative approach in different studies3–5. The instrument is highly complementary to BLI and SPR. Its main benefit is for DNA/RNA Binding Proteins, screening and ranking of small molecule inducing conformational changes (96 and 384 well plates) and enzymatic activities including polymerases, ligases and nucleases. A standard kinetics experiment requires about 100 µg of the ligand and 600 µL of the analyte at a concentration of 10 times the expected KD. We usually reserve a week for a new project.
References
1. Knezevic, J. et al. Quantitation of affinity, avidity, and binding kinetics of protein analytes with a dynamically switchable biosurface. J Am Chem Soc 134, 15225–15228 (2012).
2. Müller-Landau, H. & Varela, P. F. Standard operation procedure for switchSENSE DRX systems. European Biophysics Journal 50, 389–400 (2021).
3. Velours, C. et al. Macromolecular interactions in vitro, comparing classical and novel approaches. European Biophysics Journal 50, 313–330 (2021).
4. Marcelot, A. et al. Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners. Nucleic Acids Res 49, 3841–3855 (2021).
5. Nemoz, C. et al. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat Struct Mol Biol 25, 971–980 (2018).